If you believe that medical imaging and deep learning is just about segmentation, this article is here to prove you wrong. We will cover a few basic applications of deep neural networks in Magnetic Resonance Imaging (MRI).
The motivation is simple yet important: First, many image diagnosis tasks require the initial search to identify abnormalities, quantify measurement and change over time. Secondly, deep learning methods are increasingly used to improve clinical practice. In the field of MRI, deep learning has seen applications at every step of entire workflows. To provide some additional context, we can divide the aspects of deep learning in MRI into two parts, as in [1]:
-
the signal processing chain, which is close to the physics of MRI, including image reconstruction, restoration, and image registration, and
-
the use of deep learning in MR reconstructed images, such as medical image segmentation, super-resolution, medical image synthesis.
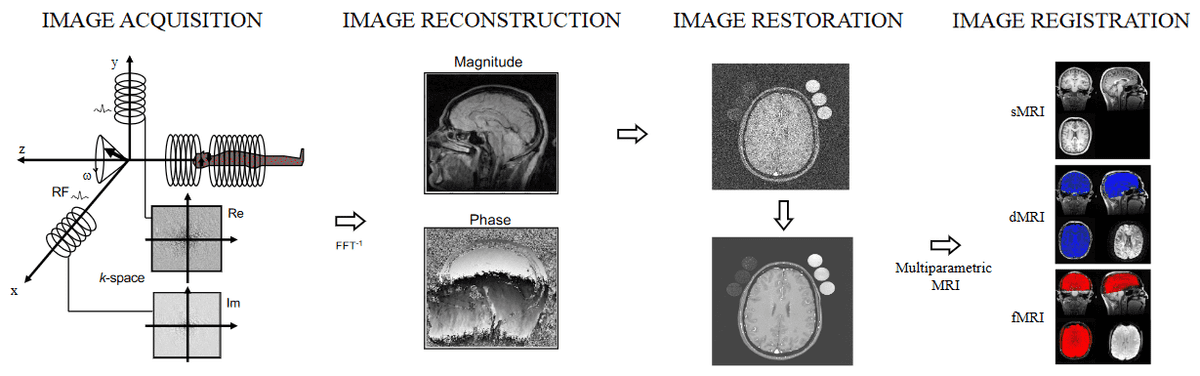
Aspects of Deep Learning applications in the signal processing chain of MRI, taken from Selvikvåg Lundervold et al. [1]
Our aim is to provide the reader with an overview of how deep learning can improve MR imaging. Before we begin, and since we are focusing on MRI, let’s clarify some concepts. This video is a great place to start, or revise, the MRI fundamentals.
For a hands-on course on AI for Medicine, check out this great course.
Medical Image Reconstruction in MRI
What is Medical Image Reconstruction: Prerequisites and Background of MRI?
The MR image generation can be quickly summed up in the following steps:
-
The MRI machine emits a radio frequency (RF) pulse at a specific frequency.
-
Radiofrequency coils send the pulse to the area of the body to be examined.
-
Then, the RF pulse is absorbed by protons, causing their redirection with respect to the primary magnetic field to change.
-
When the RF pulse is turned off, the protons “relax” back to the initial alignment by emitting radio-waves in the process.
-
Finally, the spatial information is encoded as measured data during the acquisition in the frequency domain.
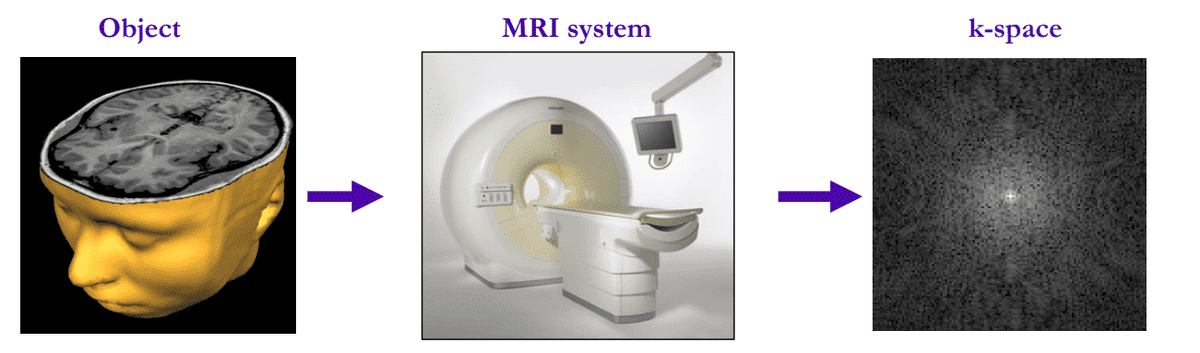
Overview of MRI measured data. Source:King’s College London
In the MRI world, they usually refer to the initial encoded acquired data as k-space. They are basically Fourier-transformed data. To go back to spatial information, we simply apply the inverse Fourier transform to obtain the MR image. This process is exactly the definition of MRI reconstruction. If you look at the 3D volume from the axial view (imagine being above the patient and looking down) it looks like this:
It’s difficult to imagine it but the k-space shown above contains equivalent information with an MR 2D slice!
Medical Image Reconstruction with deep learning
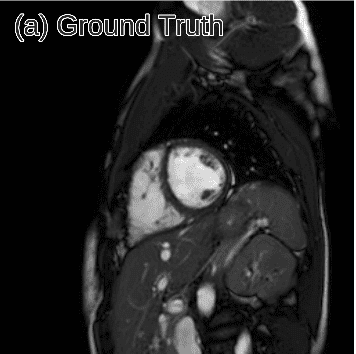
One of the first works that employed deep learning in the reconstruction process was by Schlemper et al. 2017 [2]. The authors proposed a framework for reconstructing dynamic sequences of 2D cardiac magnetic resonance (MR) images from under-sampled acquisition data, using a deep cascade of convolutional neural networks (CNNs). Their aim was to accelerate the data MRI acquisition process. It is worth noting that each 2D image frame was reconstructed independently (not optimal approach).
Interestingly, the proposed deep learning architecture method outperformed 2D compression-based approaches in terms of reconstruction error and reconstruction speed. Finally, the authors showed that their method outperforms state-of-the-art methods and can preserve the anatomical structure.
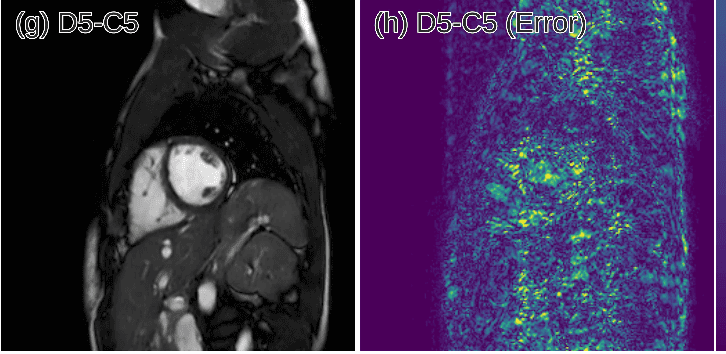
Predicted reconstruction and relative error VS ground truth
The fastMRI project: Accelerating MR Imaging with AI
Recently, Facebook AI Research (FAIR) and NYU Langone Health created a project called fastMRI. The goal is to exploit AI to speed up MRI scans, up to 10 times faster. And to achieve awesome stuff with deep learning in any domain, first you need data!
To this end, they introduced the fastMRI dataset to enable Machine Learning-based breakthroughs in the reconstruction of accelerated MR images. The raw MRI data they provide, include 8344 volumes, consisting of 167,375 slices. Moreover, they released processed MR images in DICOM format from 20,000 knee and brain examinations. That is more than 1.57 million slices for heavy deep learning. The dataset can be found here.
The main data are listed below:
-
Raw multi-coil k-space data: unprocessed complex-valued multi-coil MR measurements.
-
Ground-truth images: real-valued reconstructed images from fully-sampled multi-coil acquisitions. These may be used as references to evaluate the quality of reconstructions.
-
DICOM images: spatially-resolved images for which the raw data were discarded during the acquisition process. These images are provided to represent a larger variety of machines and settings that are present in the raw data.
However, to give you a brief idea let’s shortly discuss the proposed architecture of their recent publication [Sriram et al. 2020]:
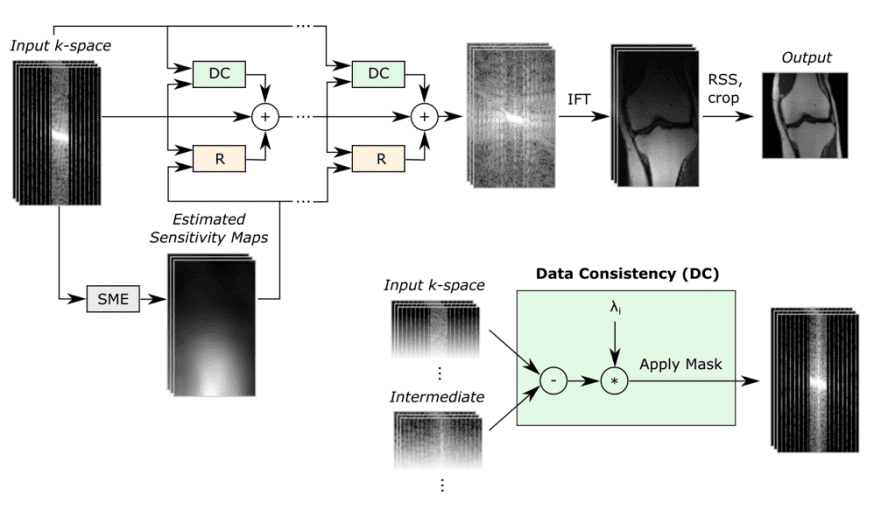
A block diagram of the reconstruction model.
The reconstruction model takes an under-sampled k-space as input and applies several cascade models (Unet-based models), followed by an inverse Fourier transform and a root-sum-squares transform.
Based on the original authors: “The Data Consistency (DC) module computes a correction map that brings the intermediate k-space closer to the measured k-space values. The Refinement (R) module maps multi-coil k-space data into one image, applies a U-Net, and then back to multi-coil k-space data. The Sensitivity Map Estimation (SME) module estimates the sensitivity maps used in the Refinement module.” ~ Sriram et al. 2020 [5]
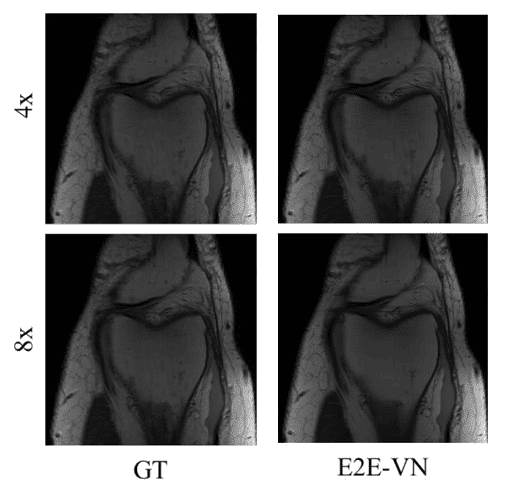
Reconstruction results with 4x and 8x the reference speed
It is impossible to analyze all the endeavors of such a huge project in a single article. Please refer to the list of publications for more info on their findings.
Medical Image Denoising and Synthesis
If you followed our GAN article-series, I am 100% sure that you know what image generation is. Image synthesis/generation is simply the learning of the distribution of the data in order to be able to produce new, realistic, crispy representative images. We can learn to produce images unconditionally, or constrain the images to satisfy a particular condition. It can be applied to medical images to solve tasks such as image denoising, image translation etc.
We will briefly describe the work proposed by Bermudez et al. 2018 [3], which was done in order to extract quantitative information from the acquired images. Their aim was to improve common image processing techniques with deep learning and provide a general framework to distinguish structural changes in the brain.
The authors used deep learning techniques to investigate implicit manifolds (latent space) of normal brains and generate new, high-quality images. This is nothing more than unconditional image generation. We start by sampling noise from a fixed distribution and try to learn a mapping to the real-world MRI data!
Further on, they also tackled image denoising with deep learning networks, which is a common processing step in MRI preprocessing. Specifically, an autoencoder with skip connections for image denoising was used, showing that the model is able to denoise medical images.
They produced T1-weighted brain MRI images using a Generative Adversarial Network (GAN) by learning from 528 examples of 2D axial slices of brain MRI. Compared to a similar model in RGB images that use thousands of images, this is an important contribution. In order to validate that the synthesized images were unique, they performed classical similarity measures (cross-correlation) with the training set.
Real and synthesized images were then assessed in a blinded manner by two imaging experts providing an image quality score of 1-5. The high-level model architecture can be illustrated below:
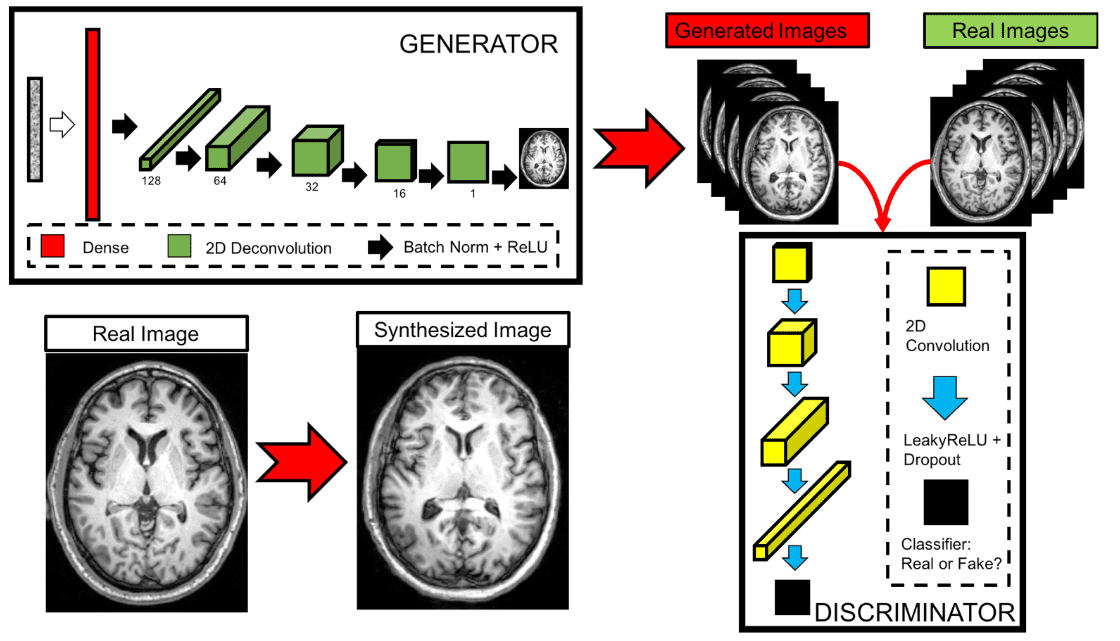
The proposed GAN architecture for MRI slices.
Main finding: the quality score of the synthetic image showed substantial overlap with that of the real images.
The radiologist’s perspective on synthetic images
Let’s examine what the medical imaging experts thought of the produced images. First, an expert radiologist mentioned that despite the comparable quality, the synthetic images were immediately given away by anatomic abnormalities. Similarly, another expert noticed brighter intensities near the center of the image compared to the boundaries in the synthetic images. These comments represent challenges in image synthesis: anatomic accuracy and signal quality. Qualitative results are illustrated below:
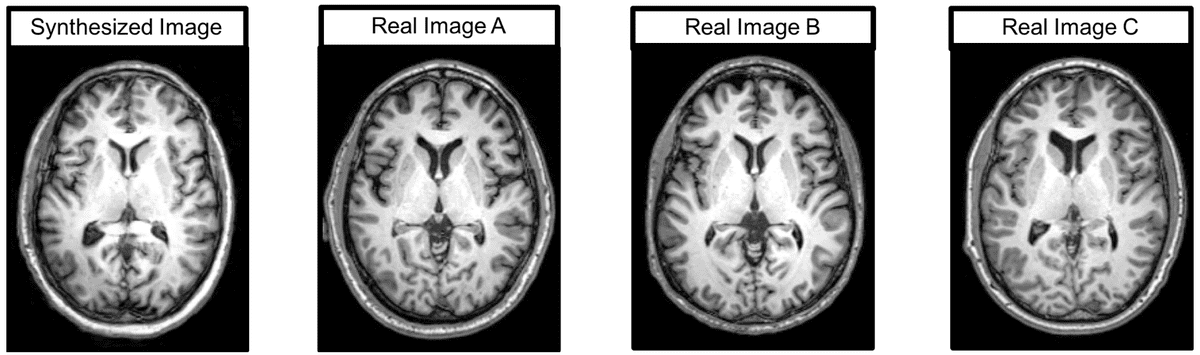
Brain MRI images, real and generated
Here is a representative synthesized image, as well as three real images with the highest correlation values.
Take-away note: the exploration of these unrealistic synthesized images may shed a light on possible structural and functional variants in brain anatomy found in healthy individuals.
Medical image translation using Cycle-GAN
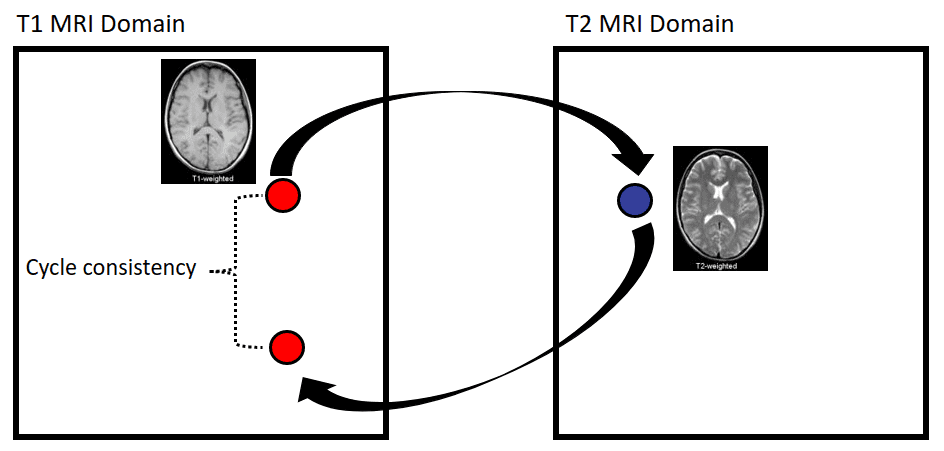
Apart from image synthesis, 2D medical image translation has been also attempted. Welander et al. 2018 [8], used Cycle GAN on brain MRI. It was one of the first works on medical image translation, specifically from T1 MRI to T2 MRI and vice versa.
Since Cycle GAN can learn to translate one domain to another and backward, it is interesting to see this concept in different medical image modalities. Briefly, instead of a single generator from T1 to T2 MRI, this model trains in parallel another generator to learn the inverse mapping from T2 to T1. Ideally, a T1 MRI that is translated to T2 and then again back to T1 through the 2 generators will result in the initial image. By constraining an image in this “cyclic” manner, we ask the model to learn a more realistic distribution.
Here are the official paper results:
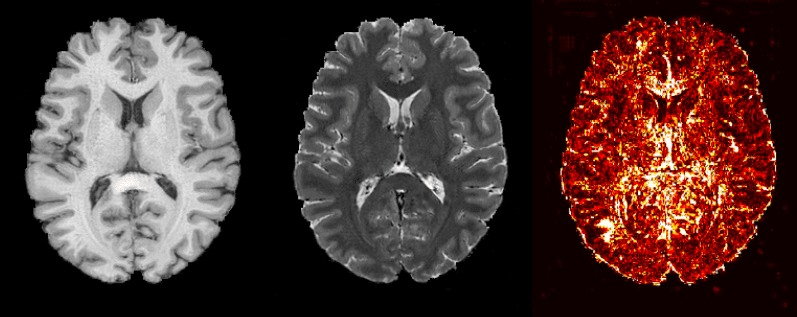
Cycle GAN’s results on medical image translation, taken from Welander et al. 2018 [8]
However, the model was trained with 2D axial slices of MRI images. The main reason was computation complexity.
Later on, in the 3D domain, Cirillo et al. [7] proposed a 3D variant of Pix2Pix GAN for multi-modal brain tumor segmentation. This time, the Generator should produce a realistic segmentation, as shown in the figure below. They further punish the model predictions with the adversarial loss. The generator is actually a 3D Unet model.
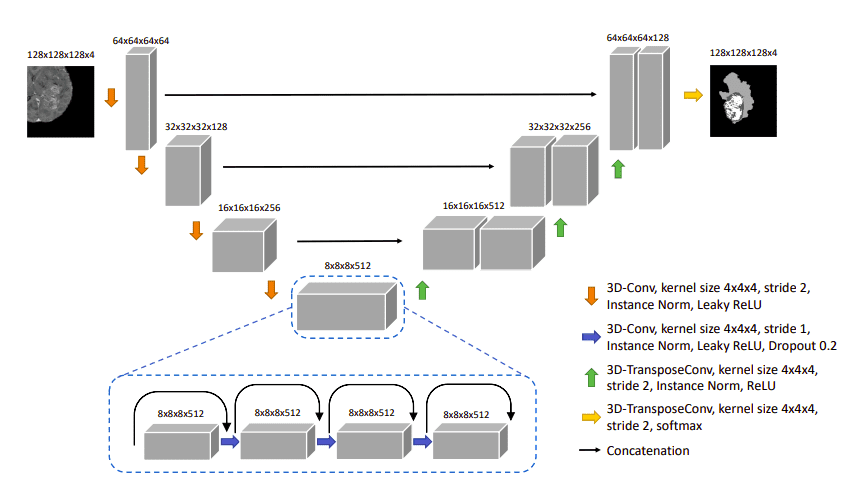
The Vox2Vox generator is a 3D Unet model.
In general, GANs are a very promising direction in medical imaging. For a thorough review on GANs in medical imaging, you can consult a review from Xin Yi et al. 2018.
Super-resolution in medical images
The task: Super-resolution can be regarded as the challenging task of estimating a high-resolution image from its low-resolution counterpart.
But how can one create such data to formulate the problem in terms of machine learning? Here is the answer:
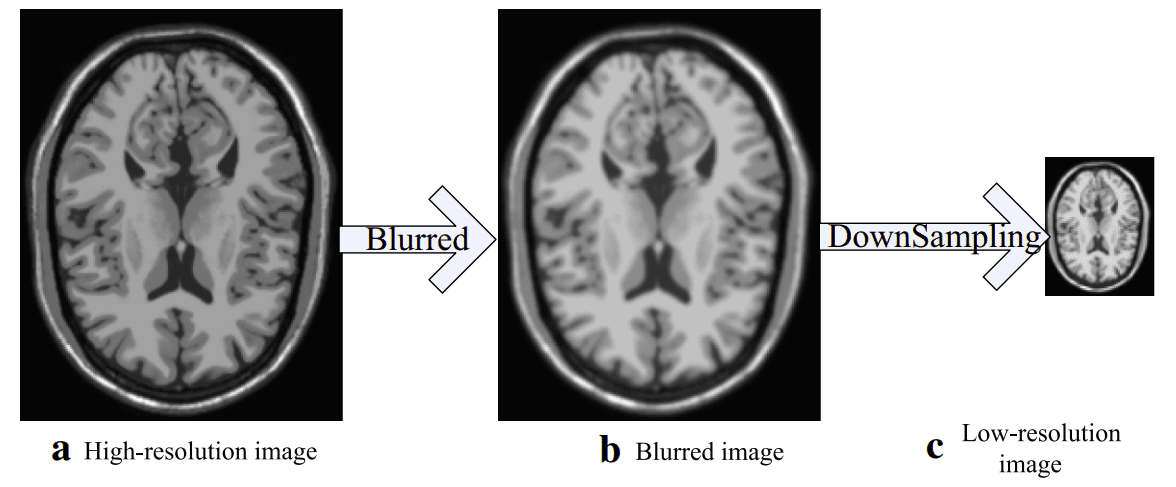
How can you create super-resolution data? Taken from [4]
You simply need to add some noise and downsample your initial image, which will be used as the ground truth.
A significant work in medical image super-resolution is performed by Liu et al. 2018 [4]. The proposed network is able to learn an end-to-end mapping from low-resolution images. To do so, they employed a multi-scale approach. As a result, the network can extract multi-scale information to recover detailed information and accelerate the convergence speed. They illustrated that fusing different paths was beneficial for recovering detailed information from a low-resolution image. It is also important that they used fully convolutional units. The trained model exhibits a reasonable performance in MRI reconstruction.
They found that a larger kernel size, an increased number of kernels, and a deeper structure, are beneficial for improving the reconstruction performance. Large kernels are closely related to the receptive field of the network. However, these features increase the computational burden and converge more slowly. Considering the ideal trade-off between performance and speed, the adopted model structure has achieved good performance.
GANs have also been proposed for medical image super-resolution. In [9], the authors trained a GAN to generate high-resolution MRI scans from low-resolution images. The architecture, which is based on the SRGAN model, adopts 3D convolutions to exploit volumetric information. Let’s see some results right away:
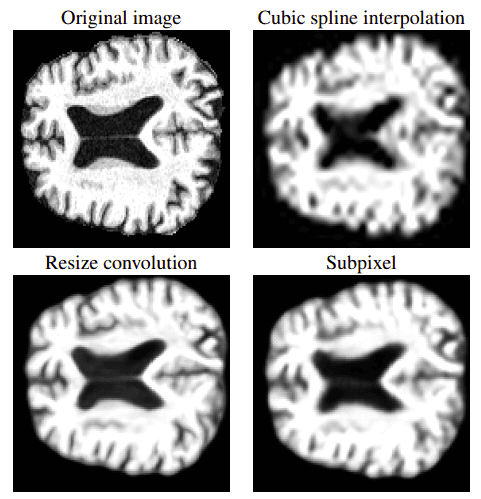
In the first row it is the original image with a common interpolation method. Below you can see the results based on the 3D SRGAN. Image is taken from [9]
Medical Image Registration
There has been increasing interest in aligning information across different medical images. Clinical applications include disease monitoring, treatment planning, etc. But what exactly is image registration?
Formally, image registration is the process of transforming images into a common coordinate system.
In terms of computer vision, I tend to think that the medical images should be aligned, so as to make meaningful comparisons. When we want to track the progress of a patient, this is critical. Because if registered, the corresponding image voxels/regions represent the same anatomical structures. For example, a PET image is by definition aligned with a CT image to understand functionality and structure respectively. In general, registration can be also used to obtain an anatomically normalized reference frame to compare the exams of multiple patients in a study (inter-patient alignment).
Below you can see an intuitive example of two registered images. Usually, translation and rotation are applied to align the images, called rigid registration:
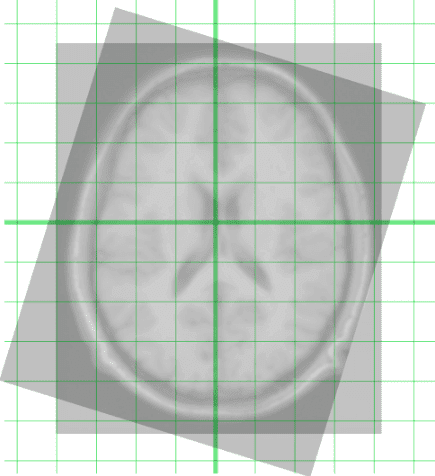
Registration of two MRI images of the brain. Taken from Wikipedia.
For machine learning, it is important to provide the model with aligned information in a multi-modal setup (intra-patient alignment). We can take advantage of the registration when building computational models of how a disease may progress.
Usually, this step is implemented using an iterative intensity-based process called Elastix [11]. Recently, a python wrapper has been developed for out-of-the-box functionality. However, many deep learning-based methods are being developed to speed up this process and provide better results.
To provide a time-scale reference, a simple registration may require roughly 2 minutes, while a trained deep neural network can perform inference in a couple of seconds.
To this end, VoxelMorph [10] is one example of how medical image registration can be performed with deep learning. Note that to optimize the model parameters a dataset of volume pairs is mandatory. Furthermore, the registration will be applicable only for the provided training modalities (i.e. Brain CT → MRI).
To achieve learning-based registration, they model a function using an architecture similar to Unet [10]. are the network parameters and f,m the fixed and moving images. The displacement field between two images and is in practice stored as an dimensional image. The registration field is computed with an identity transform and . Then, by warping the moving image m to the registration field using a spatial transformation function, they evaluate the similarity with the ground truth image f. The following image depicts a high-level overview of their approach.
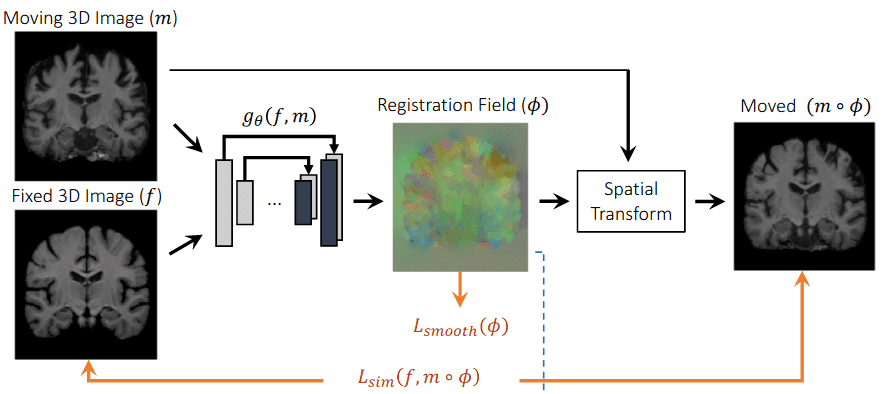
An overview of the architecture of Voxelmorph [10]. The image is taken from the original work.
For a more hands-on approach, you can try to visit their official GitHub repo. Finally, a quite famous medical image registration challenge is Learn2Reg.
Interestingly, iterative algorithms [11] still outperform deep learning approaches in medical image registration.
So, it is considered as a direction worth exploring for future research!
Conclusion
In this article, we provided the reader with a broader overview of MR tomography and deep learning. Our aim was to give a general perspective. Of course, Deep learning can aid in a lot of problems in medical imaging that is not limited to the high-level processing of final MR 3D reconstructed images. Nevertheless, one has to understand that is still just a tool. The current concern lies in the generalization capability to clinical practice. It is difficult to create datasets with the diversity of the real world. Finally, we do hope that this article inspires future collaborations between biomedical engineers, deep learning specialists, and radiologists in an interdisciplinary environment. For a more detailed overview, we encourage you to read the amazing work by Lundervold et al. [1]. As a final note, I would like to recommend the AI for Medicine course offered by Coursera, which offers exactly what you need to jump into the field.
References
- Lundervold, A. S., & Lundervold, A. (2019). An overview of deep learning in medical imaging focusing on MRI. Zeitschrift für Medizinische Physik, 29(2), 102-127.
- Schlemper, J., Caballero, J., Hajnal, J. V., Price, A. N., & Rueckert, D. (2017). A deep cascade of convolutional neural networks for dynamic MR image reconstruction. IEEE transactions on Medical Imaging, 37(2), 491-503.
- Bermudez, C., Plassard, A. J., Davis, L. T., Newton, A. T., Resnick, S. M., & Landman, B. A. (2018, March). Learning implicit brain MRI manifolds with deep learning. In Medical Imaging 2018: Image Processing (Vol. 10574, p. 105741L). International Society for Optics and Photonics.
- Liu, C., Wu, X., Yu, X., Tang, Y., Zhang, J., & Zhou, J. (2018). Fusing multi-scale information in convolution network for MR image super-resolution reconstruction. Biomedical engineering online, 17(1), 114.
- Sriram, A., Zbontar, J., Murrell, T., Defazio, A., Zitnick, C. L., Yakubova, N., … & Johnson, P. (2020). End-to-End Variational Networks for Accelerated MRI Reconstruction. arXiv preprint arXiv:2004.06688.
- Yi, X., Walia, E., & Babyn, P. (2019). Generative adversarial network in medical imaging: A review. Medical image analysis, 58, 101552.
- Cirillo, M. D., Abramian, D., & Eklund, A. (2020). Vox2Vox: 3D-GAN for Brain Tumour Segmentation. arXiv preprint arXiv:2003.13653.
- Welander, P., Karlsson, S., & Eklund, A. (2018). Generative adversarial networks for image-to-image translation on multi-contrast MR images-A comparison of CycleGAN and UNIT. arXiv preprint arXiv:1806.07777.
- Sánchez, I., & Vilaplana, V. (2018). [Brain MRI super-resolution using 3D generative adversarial networks](https://arxiv.org/abs/1812.11440\). arXiv preprint arXiv:1812.11440.
- Balakrishnan, G., Zhao, A., Sabuncu, M. R., Guttag, J., & Dalca, A. V. (2019). Voxelmorph: a learning framework for deformable medical image registration. IEEE transactions on medical imaging, 38(8), 1788-1800.
- Klein, S., Staring, M., Murphy, K., Viergever, M. A., & Pluim, J. P. (2009). Elastix: a toolbox for intensity-based medical image registration. IEEE transactions on medical imaging, 29(1), 196-205.
* Disclosure: Please note that some of the links above might be affiliate links, and at no additional cost to you, we will earn a commission if you decide to make a purchase after clicking through.